Projects
-
Statistical Modelling of TCR Repertoires for Immunotherapy and Drug Delievery Systems
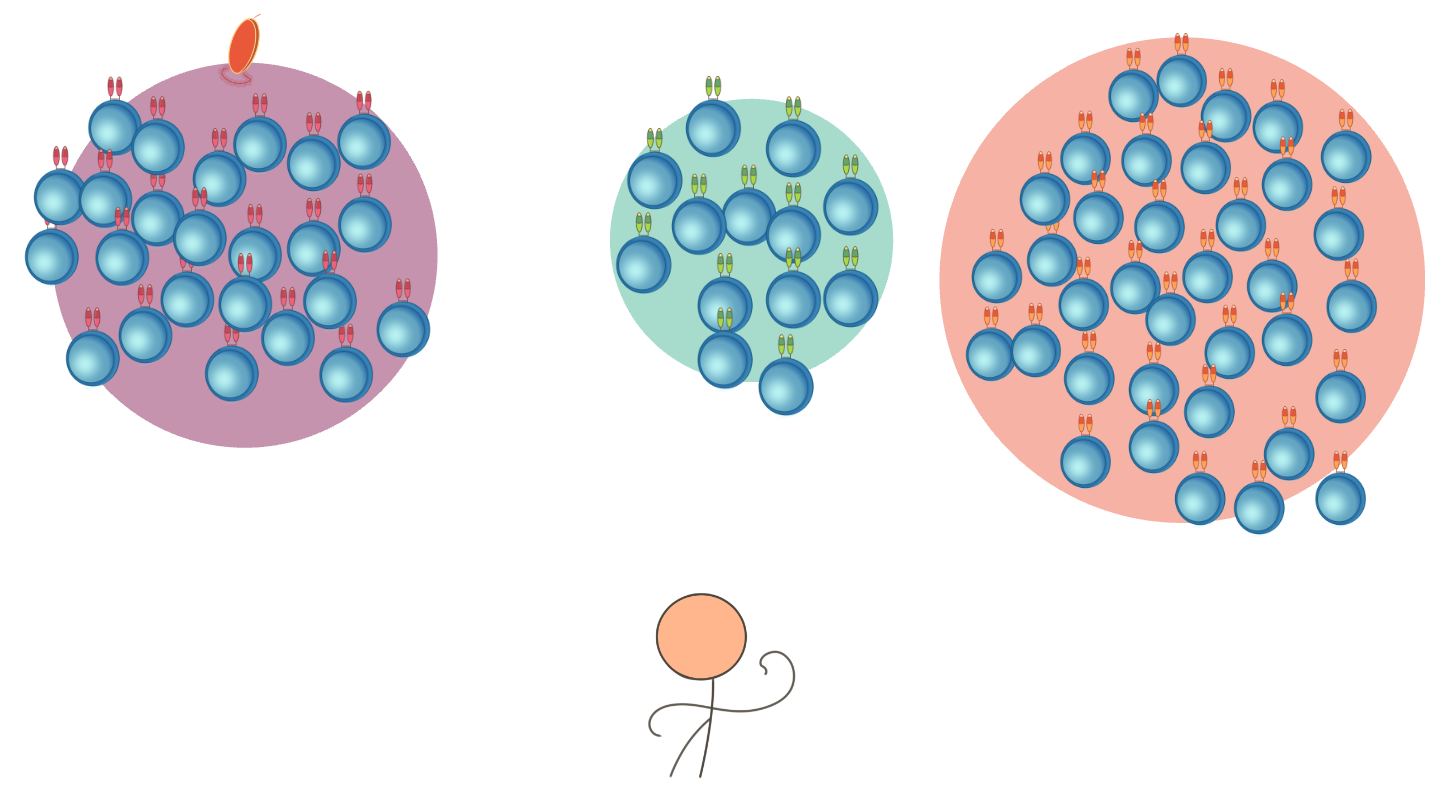
The aim of this project is disassembling mechanisms of immune system-tumour interaction, in particular to track and understand epitope spreading by means of high throughput sequencing and statistical modelling of T-cell receptor repertoires.
-
Neoepitope Prediction
Combining Deep Learning and Immunology to create a Neoepitope predictor for improving immunotherapy.